Further Calculation Functions¶
MS Peak Assignment¶
The MS assignment features of Peptide Calculator and Peptoid Calculator can be accessed through Pep-Calc API using the Mass Assignment resource.
The following example illustrates assignment of the peak list '1201.4, 1301.7, 1320.8, 1348.1, 1601.3' for the peptide sequence 'RPKPQQFFGLM'. Pep-Calc API returns a nested JSON object with the input peaks at the top level.
Top level keys point to arrays of successful assignments. A calculated mass (calcMass) is given for each assignment, alongside the deduced deletions and adducts contained within the arrays dels and mods respectively. The boolean molecular indicates whether or not an assignment corresponds to a molecular ion.
Request:
GET /peptide/mass?seq=RPKPQQFFGLM&N_term=H&C_term=OH&mz=1201.4%2C+1301.7%2C+1320.8%2C+1348.1%2C+1601.3
Response:
{
"1201.40": [
{
"calcMass": "1201.6510",
"dels": ["F"],
"mods": null,
"molecular": false
}
],
"1301.70": [
{
"calcMass": "1301.5017",
"dels": ["K", "P", "P"],
"mods": ["Na+", "Pbf"],
"molecular": false
},
{
"calcMass": "1301.5381",
"dels": ["P", "P", "Q"],
"mods": ["Na+", "Pbf"],
"molecular": false
}
],
"1320.80": null,
"1348.10": [
{
"calcMass": "1348.7194",
"dels": null,
"mods": null,
"molecular": true
}
],
"1601.30": [
{
"calcMass": "1601.7194",
"dels": null,
"mods": ["Pbf"],
"molecular": false
}
]
}
See here for a comparison with the output from Peptide Calculator’s web interface.
Beta Contiguity Calculation¶
The Beta Contiguity resource can be used to return an array of 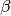 -strand propensity values for a peptide sequence, ordered by residue number. For example:
-strand propensity values for a peptide sequence, ordered by residue number. For example:
Request:
GET /peptide/betacontiguity?seq=RPKPQQFFGLM&N_term=H&C_term=OH
Response:
{
"betaContiguity": [
[1, "R", "0.0000"],
[2, "P", "0.0000"],
[3, "K", "0.0000"],
[4, "P", "1.2034"],
[5, "Q", "4.8810"],
[6, "Q", "6.0936"],
[7, "F", "7.2982"],
[8, "F", "7.2982"],
[9, "G", "4.8419"],
[10, "L", "3.6357"],
[11, "M", "3.6357"]
]
}
Please refer to the article published in the Journal of Computer-Aided Molecular Design for details of the methods used for calculation of 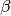 -strand propensity values.
-strand propensity values.
