
How to use Peptide Calculator
Peptide Calculator is a peptide chemical formula and molecular weight calculator and MS peak assignment tool.
Peptide sequences are entered using the standard capitalized single-letter codes. Various nonstandard multiple-letter amino acid codes are also supported and should be enclosed in parentheses, for example:
GVLYVG(pS)K(pT)KEGVVHGVA
N- and C-termini can be selected from the predefined lists. Peptide Calculator also accepts custom termini entered as chemical formulae.
MS peaks for assignment can be provided in the form of a comma-separated list and Peptide Calculator will attempt to assign them to deletion sequences and/or adducts.
A ChemDraw file is automatically generated for each peptide sequence entered, and can be downloaded via a link on the results page. It may be necessary to increase the document size in order to view the complete structure.
Where possible, Peptide Calculator will also provide estimated isoelectric point and molar extinction coefficient values for your sequence, as well as a calculated sequence difficulty profile.
Predefined termini and residues currently supported are given below (with chemical formulae). Hovering over underlined residue codes will display molecular structures.
The calculation functions of Peptide Calculator can also be used in Microsoft Excel, see the add-in page for more information.
Please send comments, questions, residue requests and bug reports to:
Theoretical isoelectric points are calculated using the bisection method algorithm described by Lukasz Kozlowski (http://isoelectric.org/).
Molar absorption coefficients are estimated using the equation described by Pace et al. (Protein Sci., 1995, 4, 2411).
Beta-strand propensity scores are calculated using an implementation of the SALSA algorithm described by Zibaee et al. (Protein Sci., 2007, 16, 906), which uses Chou-Fasman parameters (P. Y. Chou and G. D. Fasman, Biochemistry, 1974, 13, 211). Nonstandard residues are not included in sequence difficulty calculations.
Residue letter codes
1Nal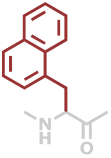 | C13H11NO |
2Nal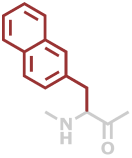 | C13H11NO |
2Pal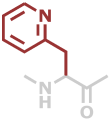 | C8H8N2O |
3Pal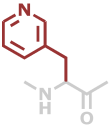 | C8H8N2O |
4Bip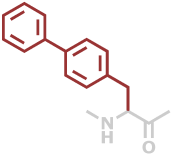 | C15H13NO |
4Bpa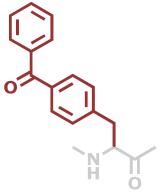 | C16H13NO2 |
A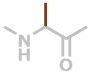 | C3H5NO |
Aad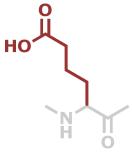 | C6H9NO3 |
Abu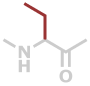 | C4H7NO |
Agl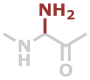 | C2H4N2O |
Ahx | C6H11NO |
Aib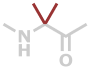 | C4H7NO |
AspOAll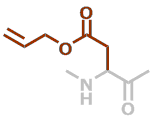 | C7H9NO3 |
Asu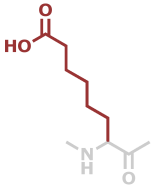 | C8H13NO3 |
Aze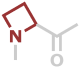 | C4H5NO |
C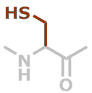 | C3H5NOS |
Cha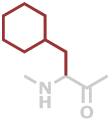 | C9H15NO |
Chg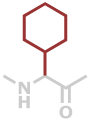 | C8H13NO |
Cit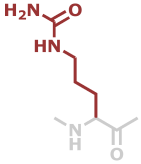 | C6H11N3O2 |
D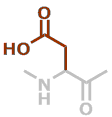 | C4H5NO3 |
Dab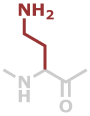 | C4H8N2O |
Dap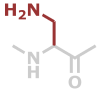 | C3H6N2O |
Dha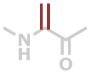 | C3H3NO |
Dhp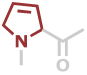 | C5H5NO |
Dip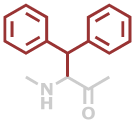 | C15H13NO |
Dpr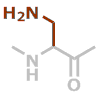 | C3H6N2O |
E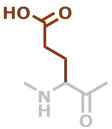 | C5H7NO3 |
F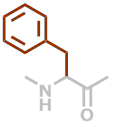 | C9H9NO |
G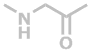 | C2H3NO |
Gla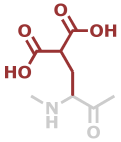 | C6H7NO5 |
GluOAll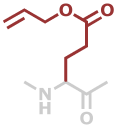 | C8H11NO3 |
H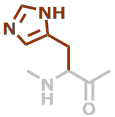 | C6H7N3O |
Har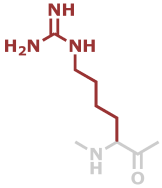 | C7H14N4O |
Hci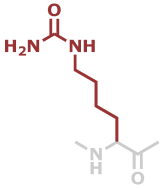 | C7H13N3O2 |
Hcy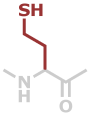 | C4H7NOS |
hCys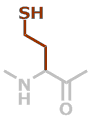 | C4H7NOS |
Hph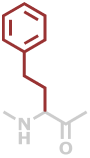 | C10H11NO |
Hpr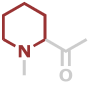 | C6H9NO |
Hyl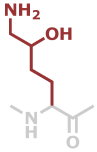 | C6H12N2O2 |
Hyp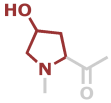 | C5H7NO2 |
I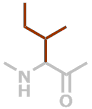 | C6H11NO |
K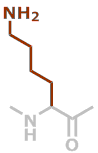 | C6H12N2O |
L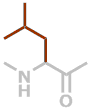 | C6H11NO |
LysAlloc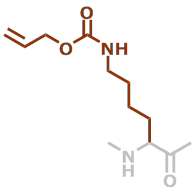 | C10H16N2O3 |
LysDde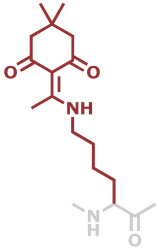 | C16H24N2O3 |
LysivDde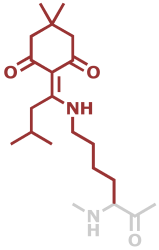 | C19H30N2O3 |
M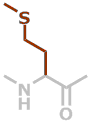 | C5H9NOS |
N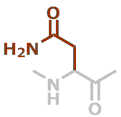 | C4H6N2O2 |
Nle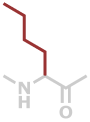 | C6H11NO |
Nva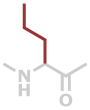 | C5H9NO |
Oic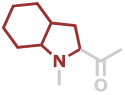 | C9H13NO |
Orn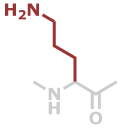 | C5H10N2O |
P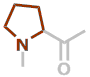 | C5H7NO |
Pal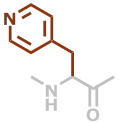 | C8H8N2O |
Pen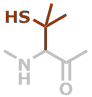 | C5H9NOS |
Phg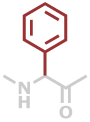 | C8H7NO |
Pip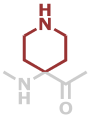 | C6H10N2O |
Pra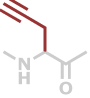 | C5H5NO |
pS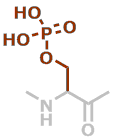 | C3H6NO5P |
pT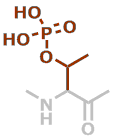 | C4H8NO5P |
pY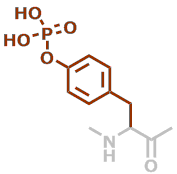 | C9H10NO5P |
Pyr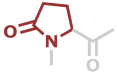 | C5H5NO2 |
Q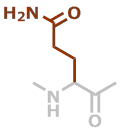 | C5H8N2O2 |
R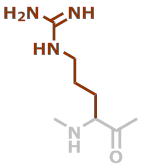 | C6H12N4O |
S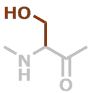 | C3H5NO2 |
T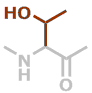 | C4H7NO2 |
Thi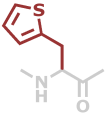 | C7H7NOS |
Tic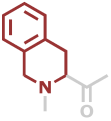 | C10H9NO |
Tle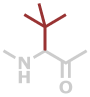 | C6H11NO |
Tpi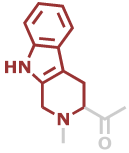 | C12H10N2O |
U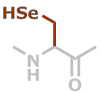 | C3H5NOSe |
V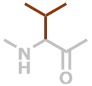 | C5H9NO |
W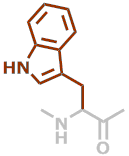 | C11H10N2O |
Y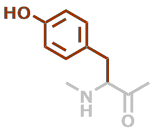 | C9H9NO2 |
Predefined N-termini
| Acetyl | CH3CO |
| Benzyl | C8H9 |
| Biotin | C10H15N2O2S |
| Fmoc | C15H11O2 |
| Methyl | CH3 |
Predefined C-termini
| Alloc Dbz | C11H12N3O3 |
| Amide | NH2 |
| Benzyl ester | C7H7O |
| Dbz | C7H8N3O |
| Ethyl 3-mercaptopropionate | C5H9O2S |
| Hydrazide | NHNH2 |
| MPAA | C8H7O2S |
| Methyl ester | OCH3 |
| N-acylurea | C8H6N3O2 |
| SEA | C4H10NS2 |
| SeEA | C4H10NSe2 |
| Sulfamyl Rink | C4H9N2O3S |