API Reference¶
Details of all available Pep-Calc API resources are listed below. All URLs are relative to http://api.pep-calc.com. Query parameters should be formatted as described on the Peptide Calculator and Peptoid Calculator help pages.
Further information about the methods used in calculations carried out by Pep-Calc API is also given on the help pages and the relevant article published in the Journal of Computer-Aided Molecular Design.
Basic Properties¶
-
GET/peptide¶
-
GET/peptoid¶ Returns basic calculated properties for the sequence, including
formulaandmolecularWeight. See Getting Started for an example output.Query Parameters: - seq – peptide/peptoid sequence string
- N_term – N-terminus chemical formula string
- C_term – C-terminus chemical formula string
Mass Assignment¶
-
GET/peptide/mass¶
-
GET/peptoid/mass¶ Returns an assignment of MS peaks passed in the
mzquery parameter. See Further Calculation Functions for an example output.Query Parameters: - seq – peptide/peptoid sequence string
- N_term – N-terminus chemical formula string
- C_term – C-terminus chemical formula string
- mz – comma-separated list of MS peaks for assignment
Ion Series¶
-
GET/peptide/ionseries¶
-
GET/peptoid/ionseries¶ Returns m/z values corresponding to the series of singly- and multiply-protonated ions calculated for both the target peptide and possible single deletions/adducts. MS peaks for assignment can optionally be passed in the
mzquery parameter.The top level array contains objects corresponding to deletion/adduct or molecular ion species. For each object the boolean
molecularindicates whether or not the species is a molecular ion, and the arraysdelsandmodsdescribe applicable deletions and adducts respectively. The arrayionscontains the series of objects corresponding to calculated m/z values for the species.For each ion the calculated m/z value (
mz) and charge state (z) are provided. The booleanassignedindicates whether any MS peaks provided have been assigned to this m/z value.Query Parameters: - seq – peptide/peptoid sequence string
- N_term – N-terminus chemical formula string
- C_term – C-terminus chemical formula string
- mz – (optional) comma-separated list of MS peaks for assignment
Isoelectric Point¶
-
GET/peptide/iso¶ Returns a calculated isoelectric point (
pI) for the peptide sequence. Anullvalue will be returned for sequences with nonexistent or out of range isoelectric points.See the Basic Properties resource for required query parameters.
Charge Summary¶
-
GET/peptide/charges¶ Returns totals of acidic (
acidicCount), basic (basicCount), uncharged (unchargedCount) and unclassified/nonstandard (otherCount) residues for the peptide sequence.See the Basic Properties resource for required query parameters.
Extinction Coefficient¶
-
GET/peptide/extinction¶ Returns calculated molar extinction coefficient values for the peptide sequence, based on either the maximum number of disulfide bonds possible (
oxidized) or the absence of disulfides (reduced).Query Parameters: - seq – peptide sequence string
ChemDraw Generation¶
-
GET/peptide/chemdraw¶
-
GET/peptoid/chemdraw¶ Returns a ChemDraw file generated for the sequence in .cdxml format.
See the Basic Properties resource for required query parameters.
Beta Contiguity¶
-
GET/peptide/betacontiguity¶ Returns an array of
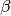 -strand propensity values for the peptide sequence, ordered by residue number. See Beta Contiguity Calculation for an example output. A
-strand propensity values for the peptide sequence, ordered by residue number. See Beta Contiguity Calculation for an example output. A nullvalue will be returned for sequences not containing any predicted beta-strand forming regions.See the Basic Properties resource for required query parameters.
